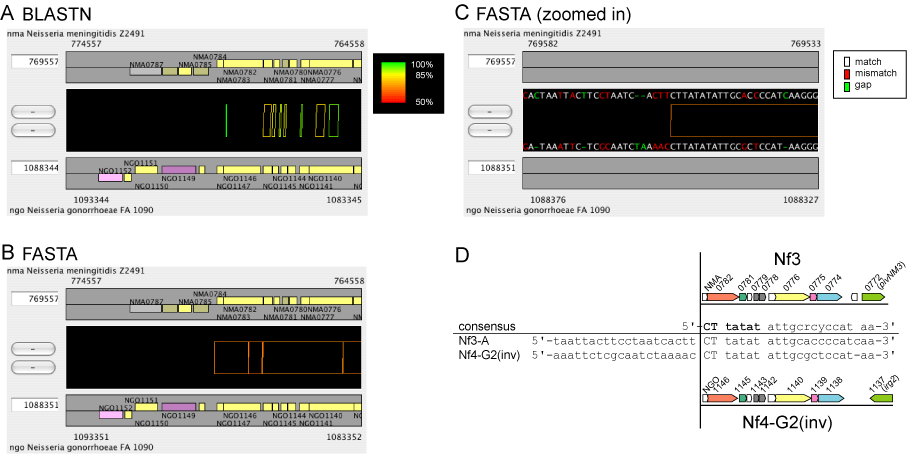
(A)-(B) Alignment of the same regions by different alignment engines. Whereas BLASTN (A) detected only partial homology between the two regions, FASTA (B), more-sensitive program than BLASTN, detected weakly similar sequences between the two regions. The regions where homology is detected by FASTA correspond to two Neisserial filamentous (Nf) phage copies, Nf3-A and Nf4-G2(inv). In (A) and (B), each alignment is colored according to percentage identity, so that 100% is colored green, 85% is colored yellow, 50% is colored red, and the remaining identities are interpolated between these colors.
(C) The actual sequence alignment in the region depicted in (B). The actual sequence alignment is displayed with colors assigned to each character as follows: white, match; red, mismatch; green, gap.
(D) Schematic representation of the phage structure. Bold face letters in the consensus sequence indicate the sequence conserved among not only these two Nf copies but also all Nf copies (not shown here). See the reference below for details.
Reference: Kawai, M., Uchiyama, I, and Kobayashi, I. (2005), Genome comparison in silico in Neisseria suggests integration of filamentous bacteriophages by their own transposase. DNA Research 12 (6), 389-401 (2005). [PubMed] [Full text]
The figure was provided by M. Kawai.