Configuring AlignmentViewer
Many properties of AlignmentViewer can be configured through property parameters. These parameters can be changed on the properties window (File ⇒ Properties). Configured properties are saved in the user home directory.
In the properties window, the properties are categorized into the following four sections.
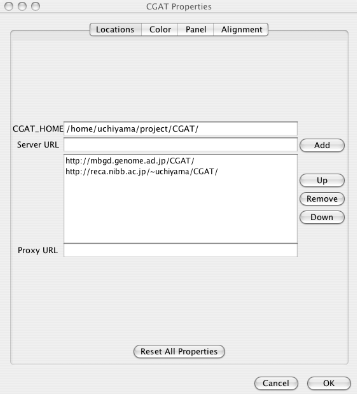
Locations: Locations of data to be loaded. See "Load data " for loading data.
CGAT_HOME: The server home directory containing the database. You can load data from the local file system, only when the CGAT database is installed in a directory accessible from the AlignmentViewer.
Server URL: Addresses of the CGAT servers. Multiple servers can be specified. You can choose which server to use when you load the data.
Proxy URL: Specify if your organization uses a proxy server. Please ask your network administrator.
Color: Color preferences. You can set two different sets of colors for the dark and light background color modes.
Background: The background color of both the alignment track and the dotplot display.
Viewing Frame: The color of the frame indicating the current scope in the dotplot display.
Match/Mismatch/Gap: Colors for displaying nucleotide sequence alignments in the annotation track.
Identity: Colors for drawing alignment regions according to the percentage identities (the identity color mode). Colors of the three points (highest, middle, and lowest percentages) can be specified, and the others are interpolated between these colors.
Gene Attribute: Colors for drawing genes in the annotation tracks using gene attribute values. Colors of the highest and lowest values can be specified, and the remaining values are interpolated between them.
Panel: Hight of the tracks and the maximum number of tracks.
Maximum # of Segment Tracks: Maximum number of feature segments to be loaded. Possibly, the height of the tracks should also be modified (lowered) to display all the information within the screen when you want to increase the number of segment tracks.
Height of Alignment/Gene/Segment Area: Hight of the tracks in the alignment display can be set individually for the alignment track, the gene annotation tracks and the feature segment tracks.
Alignment: Alignment parameters.
Match/Mismatch/Open Gap/Extension Gap: The scoring system for dynamically calculating alignments in the AlignmentViewer program.
Size of Realignment: Maximum length of the displayed region (scope) within which the re-alignment of the sequences is allowed by right click on the alignment track.