MBGD Top > Quick Tour > Select & View ortholog table
Select & View ortholog table
- MBGD provides several pre-computed ortholog tables, each of which is created using a different set of organisms.
By default, the standard ortholog table is selected, which contains one genome from each genus, covering the entire taxonmic range.
To see the global picture of the ortholog table currently selected,
click "Orthologous Table" on the top page.
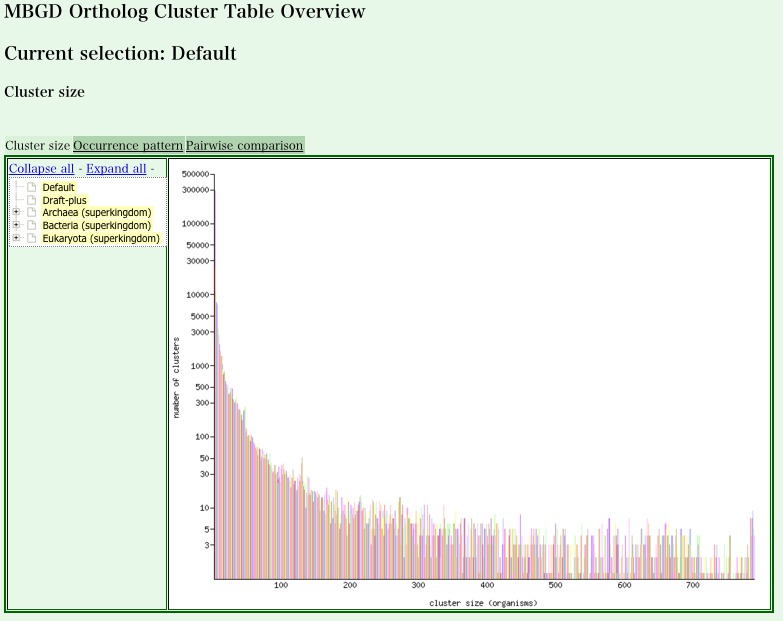
- Ortholog Cluster Table Overview page is displayed. Here, you can see a histogram showing the distribution of the cluster size (the number of organisms included) in the default ortholog table.
-
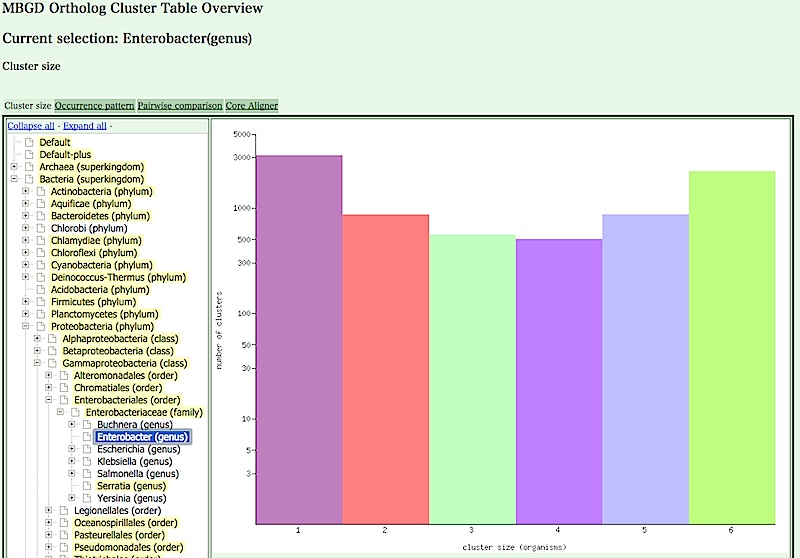
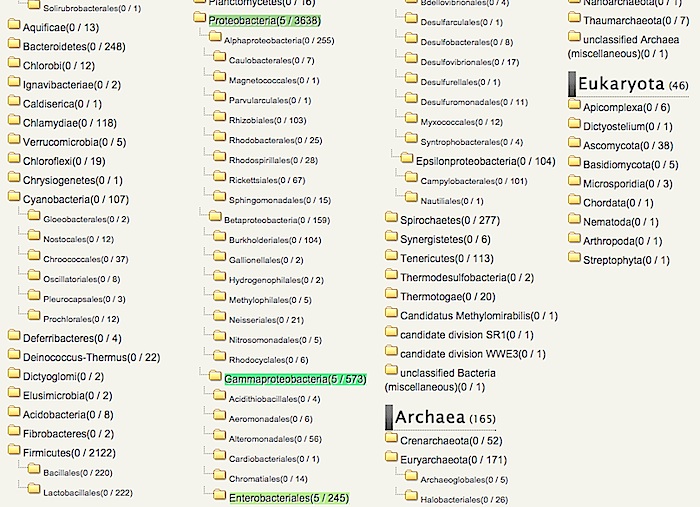
In addition to the default ortholog table, MBGD provides an ortholog table for each major taxon that contains at least six representative genomes.
You can choose a pre-calculated ortholog table from the taxonomy tree in the left side panel.
Find "Enterobacter (genus)" (located under Bacteria/Proteobacteria/Gammaproteobacteria/Enterobacteriales/Enterobacteriaceae) on this taxonomy tree, and click it.
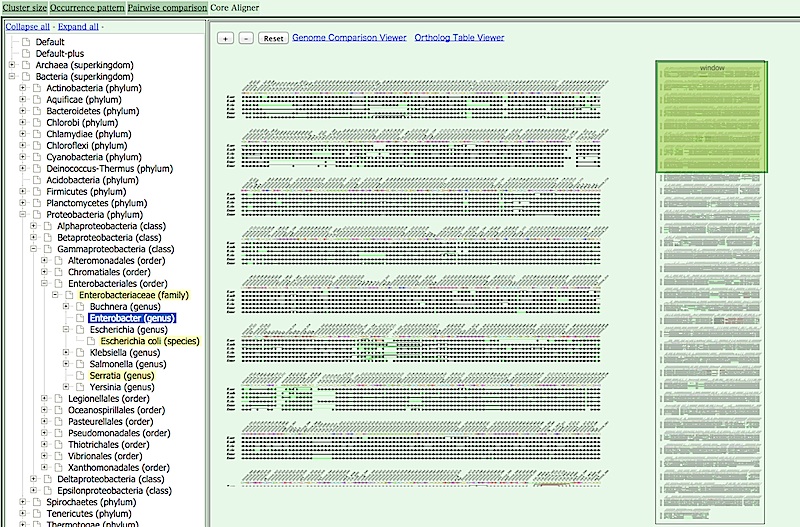
- You can change the view of the clustering result by clicking the tab at the top of the view panel.
Several views are available, including the following:
- Cluster size : Histogram of cluster size
- Occurrence pattern : Bar graph showing the relationship between occurrence pattern (representing presence or absence of the orthologs in each genome/taxon) and functional category of each orthologous group
- Pairwise comparison : Similarity matrix for pairwise genome comparisons from which one can invoke the CGAT program to display dotplot between any pair of genomes
- CoreAligner : Diagram showing the syntenically conserved core structure
- You can go back to the top page by clicking the MBGD logo at the bottom of the page. When you go back to the top page, the set of organisms
included in the currently selected cluster table is retained, and highlighted in the taxonomy table in the top page.
Let's move to the next tour: 2. Searching against an orthologous gene table.