MBGD Top > Quick Tour > MyMBGD
My MBGD: Add your own genome data to MBGD
-
My MBGD is a service that allows you to add your own
genome data to the current MBGD. To use this functionality,
you must prepare your genome data in either of the following formats:
- Combined data in GenBank format
- Protein (and genomic DNA) sequence data in Fasta format and annotation data in a tab-delimited text file (see Help for details).
To register your data, firstly you must enter the "My MBGD Mode" from the top page menu.
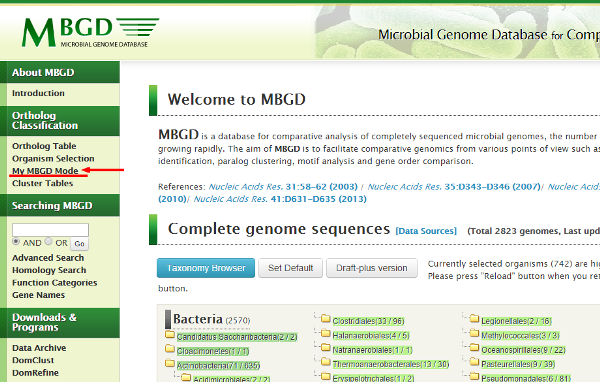
- Then the top page is redisplayed with slightly different menu items.
This is the "My MBGD Mode". You can quit this mode by clicking
"Exit My MBGD mode".
Now, click "My MBGD" to enter the data input form.
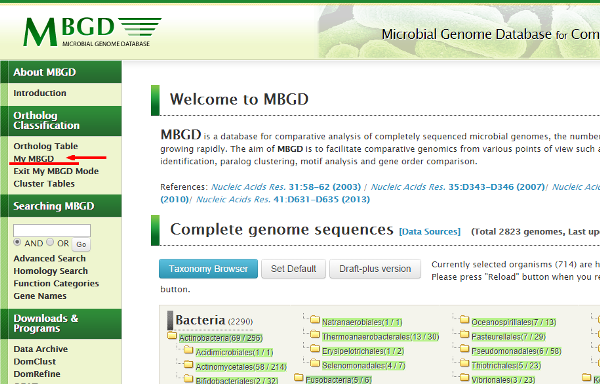
- The data input form is displayed.
In My MBGD, there are two sub mode: "guest user mode" and "registered user mode". In the guest user mode (default), MBGD maintains users' sessions using the HTTP cookie mechanism. As a result, you may lost your data if your cookie is expired (default 30 days, but we cannot guarantee this date because the cookie is actually stored in your browser). Moreover, in this mode, you cannot access the same data from different browsers. Therefore, we recommend you to create your user account by clicking the [Create Account] tab before you register your data (or if you already have an account, press the [Login] tab to log-in).
In the following example, we use the guest user mode.
- Enter the genome information.
- Species name: Carsonella ruddii
- Abbreviation Name: C. ruddii (or just press the "Set" button)
- Species code: u_cru
- Strain: PV

Now you can find the the Taxonomy ID of the strain Candidatus Carsonella PV and enter it:- Taxnomy ID: 387662
Tip: If you cannot find any appropriate taxonomy record, you can specify any taxonomy node near your organism. If you specify no taxonomy ID or a non-existing ID, your data will be treated as "Unclassified".
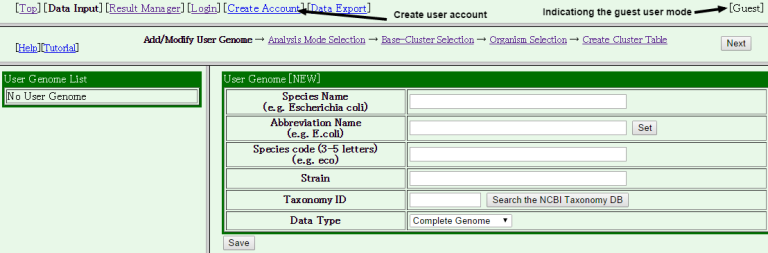
- Next, enter the chromosome information.
First, specify the type of your input file; for complete genome data, you can choose either a GenBank format file (GenBank) or a tab delimited gene file plus a fasta formatted protein sequence file (GeneTab+ProteinSeq).
Here, we choose GenBank format and press "Create New Chromosome" button.
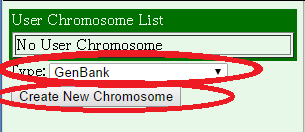
Then, enter the information. In this case, the choromosome name is automatically assigned as "chromosome 1", but you can change it if you want. If your data is in a GenBank format file that contains all required information, then you can just enter the file name into the "Data in GenBank Format" field. Otherwise, you should see Help. After you entered information, press the "Save" button below, and press "Ok".
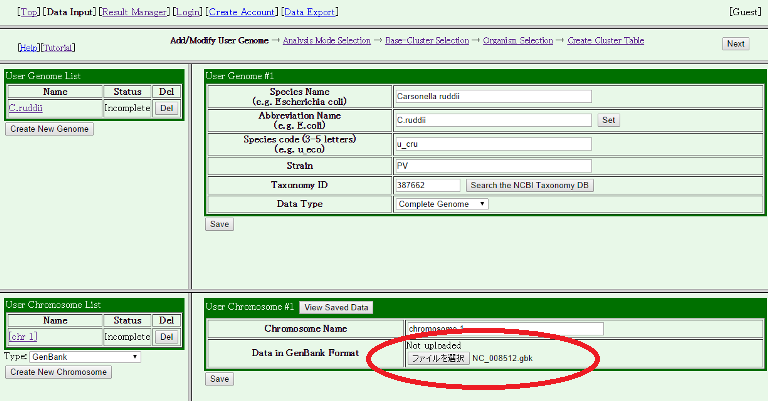
Check if the Status field in the "User Genome List" and the "User Chromsome List" tables in the light side panel are changed from "Incomplete" to "Ok." You can also check whether the gene information in your input data is correctly read, by pressing the "View Saved Data" button. If your genome has multiple chromosomes, repeat this process until all data have been uploaded.
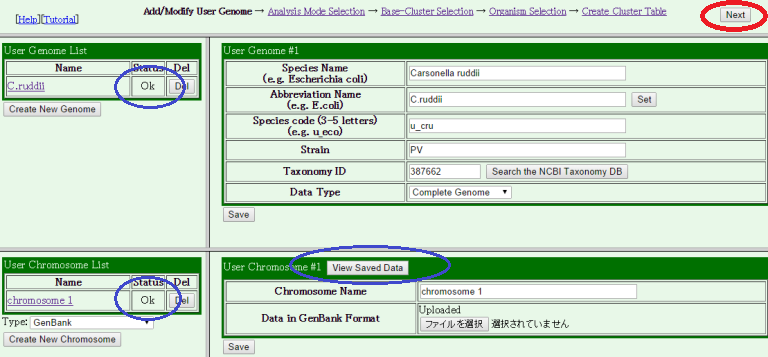
If everything is OK, then press the "Next" button at the top right corner.
- Next, choose analysis mode.
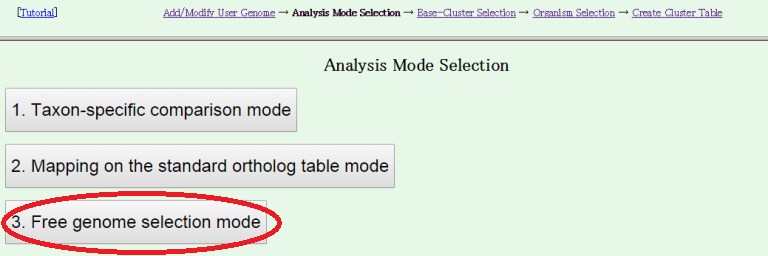
There are three analysis modes:- Taxon specific comparison mode
First choose a taxon to be analyzed and then specify a set of genomes to be compared within that taxon. In this mode, users can choose either from scratch or adding to a taxon-specific ortholog table. - Mapping on the standard ortholog table mode
Selected genomes will be added to the standard ortholog table using MergeTree. - Free genome selection mode, which is equivalent to the previous MyMBGD mode
Users can freely choose a set of genomes to compare including user genomes, complete genomes and draft genomes and conduct ortholog analysis using DomClust.
- Taxon specific comparison mode
- Next, choose genome sequences to be compared.
The maximum number of genomes to be selected is limited according
to the size of your data.
In this case, you can choose maximally 50 genomes.
Here, we choose one representative genome from each family within the class "Gammaproteobacteria". To do so, first press "Cluser All" to cancel the previous selection, search "Gammaproteobacteria(class)" and press the checkbox in this line to choose all the genomes in this taxon and then goto the page top and choose "family" in the pull-down menu and press "Select". Do not forget to select your input genome in the left side table.
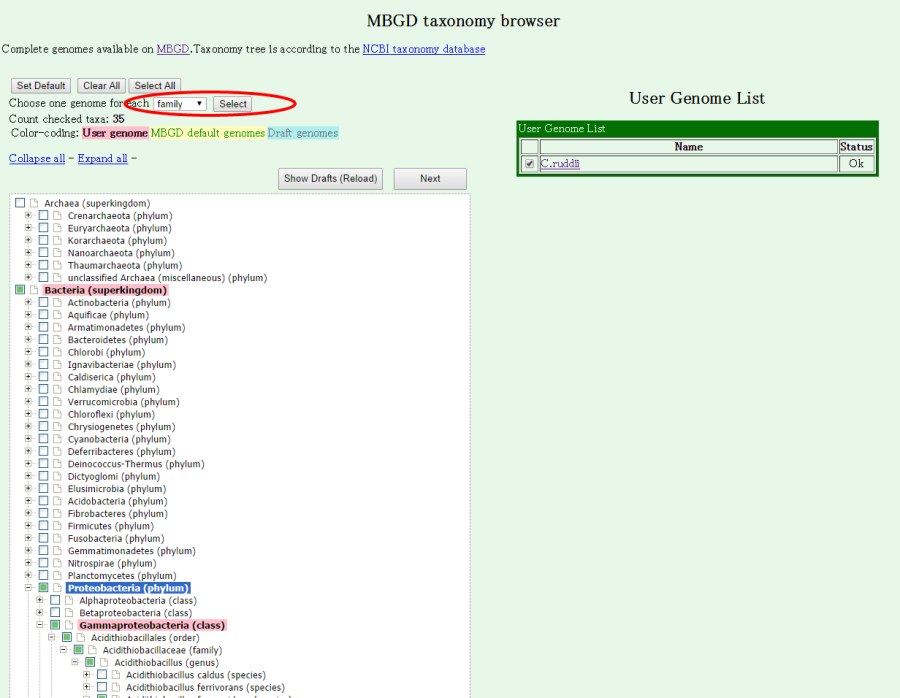
You can check the number of selected organisms in "Count checked taxa". Now, press "Next" to fix the selection and go to next.
- The Homologous Gene Table Cration page is displayed. Press "Create/View Cluster Table" to start building.
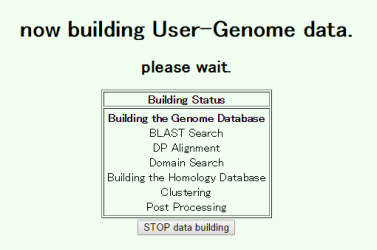
The building process includes database construction, all-all homology search, motif search and orthlog clustering. In this case with relatively small data, probably you should wait ten minutes or so until the process is completed, but it may take more time if the server is busy.
Please be patient!
- You are done! The clustering result is displayed, in which
your data is displayed with species code.
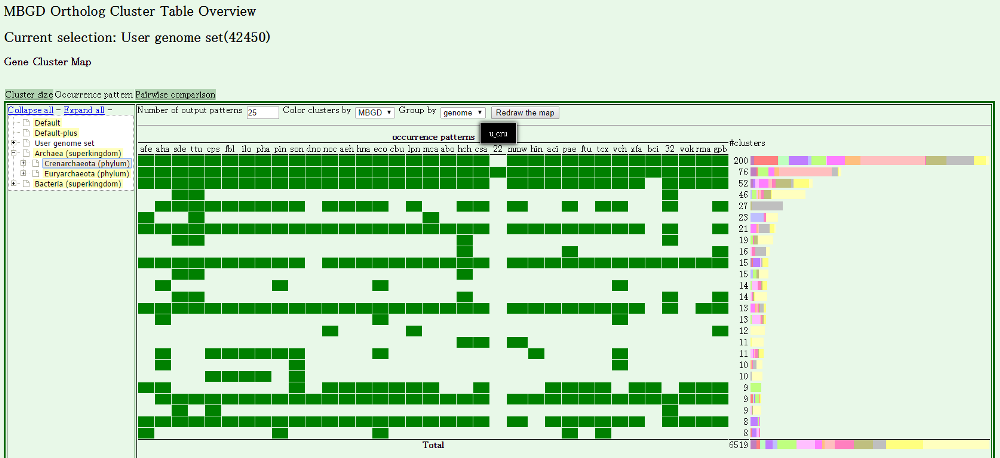
You can use every function of MBGD as in the usual mode.
- Do not forget to "Exit My MBGD mode" if you want to
return to the usual mode.
After you exit the My MBGD mode, your data will disappear. However, the data still remains on the server so that you can access the data when entering the My MBGD mode again. But please remember that, in the guest user mode, your data may be lost after your cookie record is expired.